Sample the split proportion - proportion of removed workers in a managed split
Source:R/Class-SimParamBee.R
splitPFun.RdSample the split proportion - proportion of removed workers in a
managed split - used when p = NULL - (see
SimParamBee$splitP).
This is just an example. You can provide your own functions that satisfy your needs!
splitPUnif(colony, n = 1, min = 0.2, max = 0.4)
splitPColonyStrength(colony, n = 1, nWorkersFull = 100, scale = 1)Arguments
- colony
- n
integer, number of samples
- min
numeric, lower limit for
splitPUnif- max
numeric, upper limit for
splitPUnif- nWorkersFull
numeric, average number of workers in a full/strong colony for
splitPColonyStrength(actual number can go beyond this value)- scale
numeric, scaling of numbers in
splitPColonyStrengthto avoid to narrow range when colonies have a large number of bees (in that case changenWorkersFulltoo!)
Value
numeric, split proportion
Details
splitPUnif samples from a uniform distribution between values
0.2 and 0.4 irrespective of colony strength.
splitPColonyStrength samples from a beta distribution with mean
a / (a + b), where a = nWorkers + nWorkersFull and b =
nWorkers. This beta sampling mimics larger splits for strong colonies and
smaller splits for weak colonies - see examples. This is just an example -
adapt to your needs!
The nWorkersFull default value used in this function is geared
towards a situation where we simulate ~100 workers per colony (down-scaled
simulation for efficiency). If you simulate more workers, you should change
the default accordingly.
Functions
splitPColonyStrength(): Sample the split proportion - the proportion of removed workers in a managed split based on the colony strength
See also
SimParamBee field splitP
Examples
splitPUnif()
#> [1] 0.2833048
splitPUnif()
#> [1] 0.2040136
p <- splitPUnif(n = 1000)
hist(p, breaks = seq(from = 0, to = 1, by = 0.01), xlim = c(0, 1))
 # Example for splitPColonyStrength()
founderGenomes <- quickHaplo(nInd = 2, nChr = 1, segSites = 100)
SP <- SimParamBee$new(founderGenomes)
SP$nThreads = 1L
basePop <- createVirginQueens(founderGenomes)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
drones <- createDrones(x = basePop[1], nInd = 15)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony <- createColony(x = basePop[2])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony <- cross(colony, drones = drones)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony <- addWorkers(colony, nInd = 10)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
nWorkers(colony) # weak colony
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
splitPColonyStrength(colony)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
splitPColonyStrength(colony)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony <- addWorkers(colony, nInd = 100)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
nWorkers(colony) # strong colony
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
splitPColonyStrength(colony)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
splitPColonyStrength(colony)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
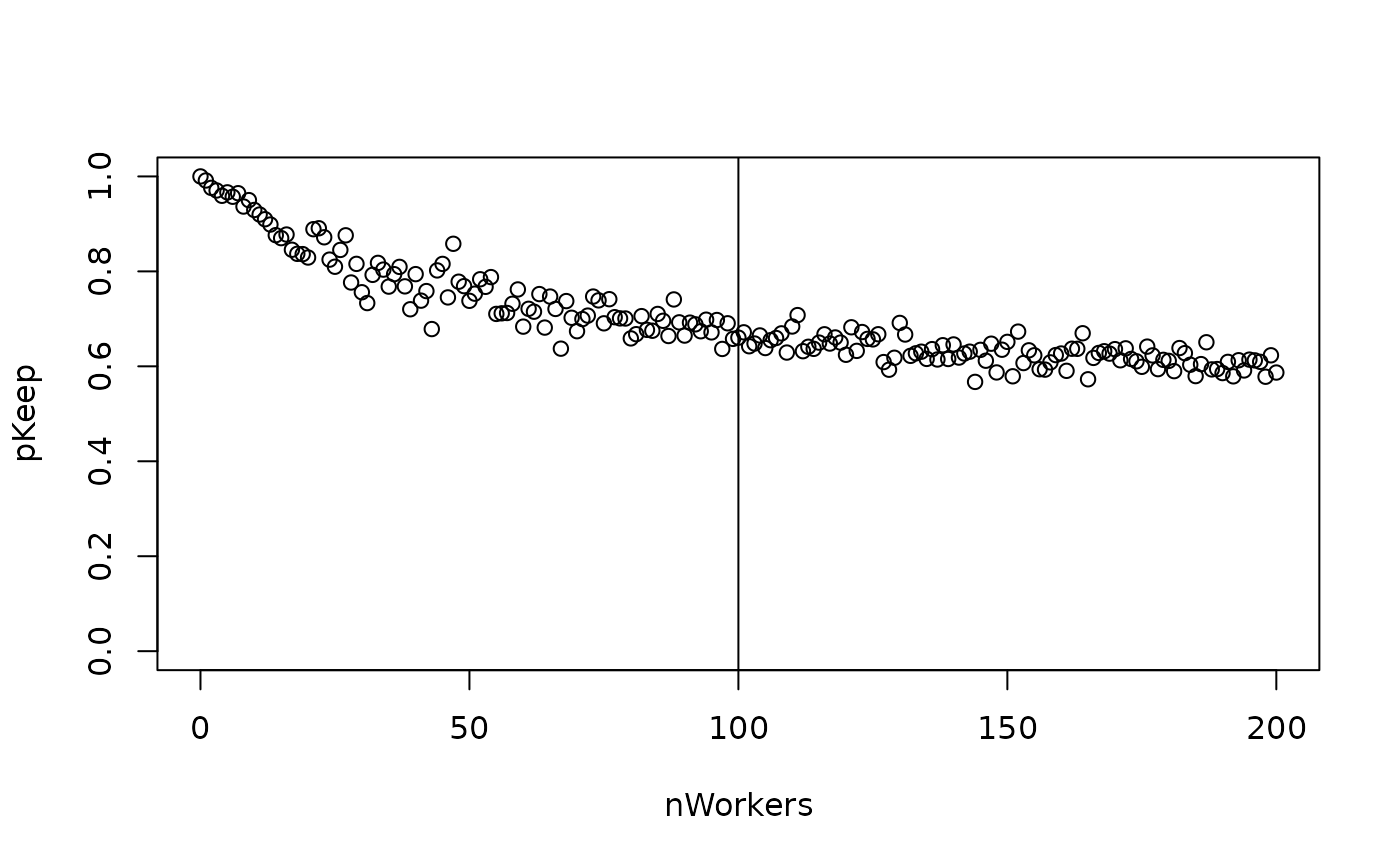
# Logic behind splitPColonyStrength()
nWorkersFull <- 100
nWorkers <- 0:200
splitP <- 1 - rbeta(
n = length(nWorkers),
shape1 = nWorkers + nWorkersFull,
shape2 = nWorkers
)
plot(splitP ~ nWorkers, ylim = c(0, 1))
abline(v = nWorkersFull)
# Example for splitPColonyStrength()
founderGenomes <- quickHaplo(nInd = 2, nChr = 1, segSites = 100)
SP <- SimParamBee$new(founderGenomes)
SP$nThreads = 1L
basePop <- createVirginQueens(founderGenomes)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
drones <- createDrones(x = basePop[1], nInd = 15)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony <- createColony(x = basePop[2])
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony <- cross(colony, drones = drones)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony <- addWorkers(colony, nInd = 10)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
nWorkers(colony) # weak colony
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
splitPColonyStrength(colony)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
splitPColonyStrength(colony)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
colony <- addWorkers(colony, nInd = 100)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
nWorkers(colony) # strong colony
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
splitPColonyStrength(colony)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
splitPColonyStrength(colony)
#> Error in get(x = "SP", envir = .GlobalEnv): object 'SP' not found
# Logic behind splitPColonyStrength()
nWorkersFull <- 100
nWorkers <- 0:200
splitP <- 1 - rbeta(
n = length(nWorkers),
shape1 = nWorkers + nWorkersFull,
shape2 = nWorkers
)
plot(splitP ~ nWorkers, ylim = c(0, 1))
abline(v = nWorkersFull)
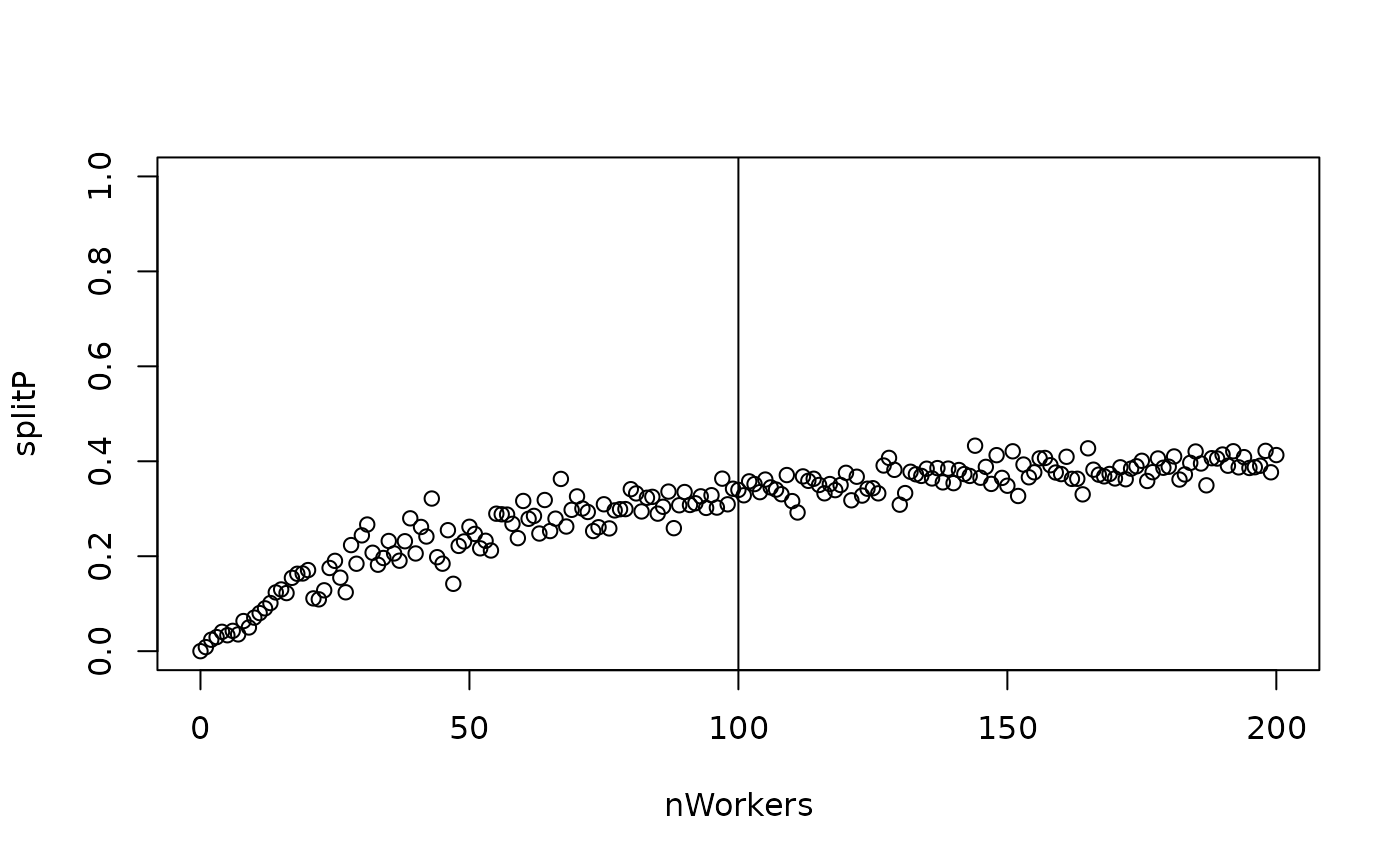 pKeep <- 1 - splitP
plot(pKeep ~ nWorkers, ylim = c(0, 1))
abline(v = nWorkersFull)
pKeep <- 1 - splitP
plot(pKeep ~ nWorkers, ylim = c(0, 1))
abline(v = nWorkersFull)